Most DNA-based methods for intestinal microbiota characterisation rely on PCR amplification with broad-range primers at an early stage of the protocol. Even though reference is frequently made to ‘universal primers’ capturing all bacteria, strictly speaking such oligonucleotides do not exist. Therefore, general bacterial analysis based on 16S rDNA sequencing is in most cases at least slightly biased, and in the worst case (primers with poor coverage) it is biased to the extent that no conclusions can be drawn on the composition of the microbial community analysed.
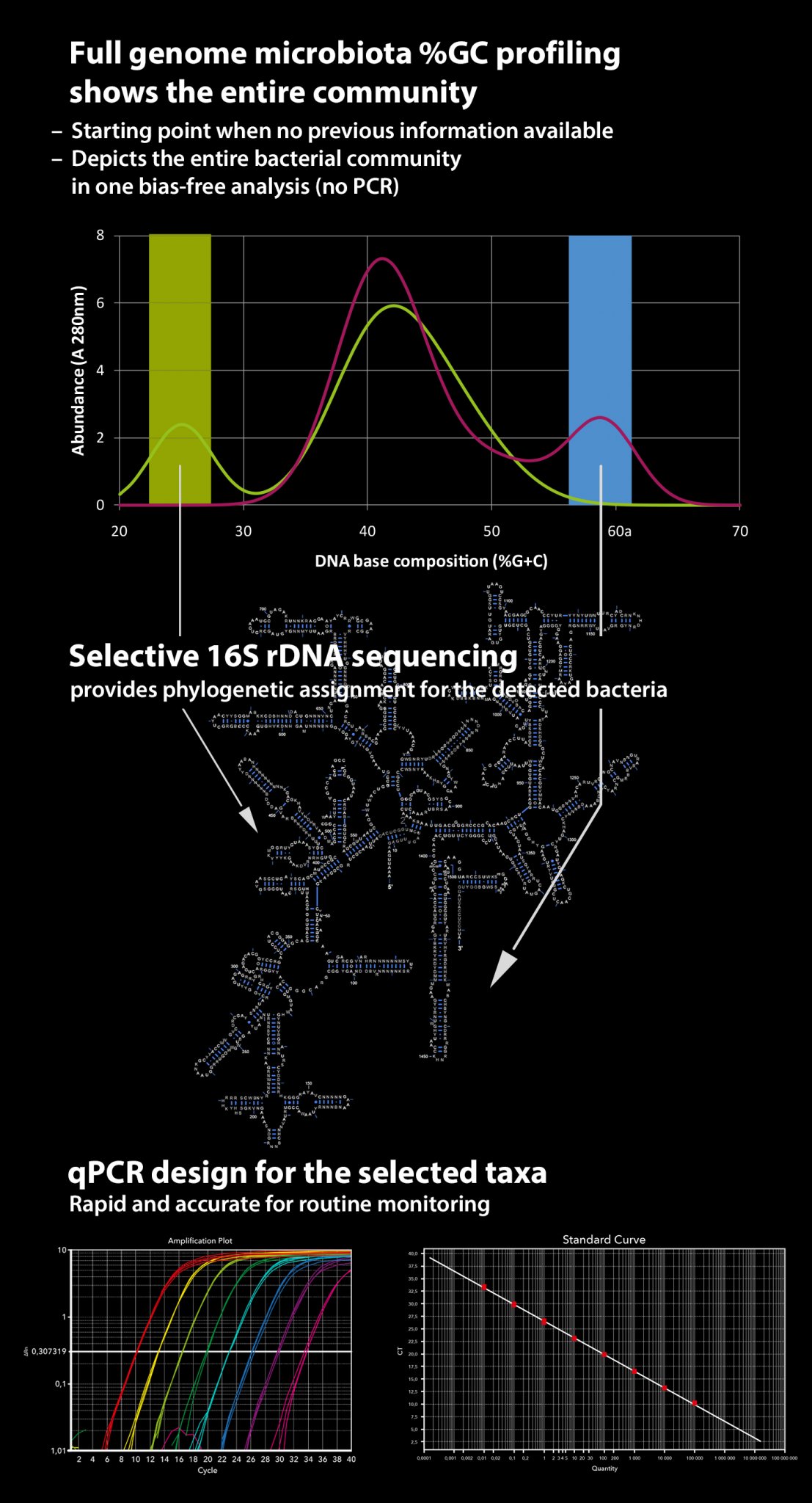
Percentage guanine + cytosine (%G+C) profiling is a method that physically fractionates bacterial chromosomes based on their G+C content. Unlike methods depending on PCR amplification, %G+C profiling remains independent of primers and probes as well as any other a priori knowledge of microbiota composition. Hence, it quantitatively and accurately illustrates the profile of the bacterial community, including all bacterial residents present.
As a stand-alone method, %G+C profiling does not provide definite taxonomic names for the bacteria present. However, it reveals taxonomically relevant characteristics of the microbiota, which facilitates subsequent more detailed analyses, such as 16S rDNA sequencing. A great advantage is that the sequencing can be targeted to the specific %G+C fractions of interest to reveal the identity of the bacteria, e.g. those that respond to an experimental diet or that are characteristic of enteric disease. In addition, when 16S rDNA sequencing is applied to an individual %G+C fraction representing chromosomes of only a small number of bacterial species, PCR-related biases due to primer mismatching, differences in G+C percentages and chimeric sequence artefacts are minimised.
Alimetrics has applied the combination of %G+C profiling and 16S rDNA sequencing to numerous applications such as gastrointestinal tract microbiota of humans, monogastric production and companion animals, ruminants, environmental samples and process samples. Even in comparative studies where other DNA-based techniques have failed to detect significant differences in microbiota composition, the combined technique has revealed the underlying bacterial shifts with high relevance to the application area.